Running Jobs
Running jobs on any HPCC server
All jobs at HPCC must:
- start from dedicated file system called scratch.
- use dedicated job submission system (job scheduler) .
- all user data (home directories) are kept in separate file system called /global/u.
Servers in free and advanced tier (Blue Moon, Penzias, CRYO and Appel) are attached via 40 Gbps interconnect to /scratch and /global/u (called DSMS) file systems. The former is a small disk based parallel file system mounted on all nodes (compute and login) and the latter is large, slower file system (holding all users' home directories /global/u/<userid>) mounted only on login node(s). Both file systems have moderate bandwidth of several hundred MB per second. Every home directory for free and advanced tier servers has a quote of 50GB. The latter can be expanded by submitting motivated request to HPCC. bal/u file s file system is backup-ed with retention time of backup 30 days. Because atch is mou is mounted on all compute nodes all jobs on any server must start from scratch only. Jobs ot be started from u from user's home: /global/u/<userid>
Arrow server and condo servers are attached to a separate hybrid (NVMI + hard disks) fast file system called HPFFS which can provide speeds of 25-30Gb/s write and 45-50 GB/s read. The /scratch and /global/u are part of the same HPFFS file system, but scratch is optimized for predominant access of the fast NVMI tier of HPFFS. All jobs must start from scratch. Jobs cannot be started from user's home: /global/u/<userid> It is important to mention that data on /global/u on HPFFS file system are not backup-ed since this equipment is not integrated in HPCC infrastructure. Every user home directory has a quote of 100 GB. The latter can be expanded by submitting motivated request to HPCC.
Before submitting any job the users must prepare/move/copy data into their /scratch/<userid> directory. Users who submit jobs on free and advanced tier machines can transfer data to /scratch/<userid> using the file transfer node (cea) or GlobusOnline. HPCC recommends a transfer to user's home directory first ( /global/u/<userid> ) before copy the needed files from user's home directory to /scratch/<userid> . In addition, cea and Globus online allows the transfer of user's files directly to /global/u/<userid>. The input data, job scripts and parameter(s) files can be locally generated with use of Unix/Linux text editor such as Vi/Vim, Edit, Pico or Nano. MS Windows Word is a word processing system and cannot be used to create job submission scripts.
Running jobs on Arrow
Arrow's File system
Arrow is attached to NSF funded 2 PB global hybrid file system. The latter holds both users' home directories (/global/u/<userid> ) and users' scratch directories (/scratch/<userid>). The underlying files system manipulates the placement of the files to ensure the best possible performance for different file types. It is important to remember that only scratch directories are visible on nodes. Consequently jobs can be submitted only from /scratch/<userid>. directory. Users must preserve valuable files (data, executables, parameters etc) in /global/u/<userid>. It is impotant to remember that 2Pb PFSS file system is not connected to main infrastructure and thus users cannot use the file transfer node (CEA) or GLOBUS online to move files from/to PFSS.
Copy files from/to Arrow
Because Arrow is detached from main HPC infrastructure the user files can only be tunneled to Arrow with use of ssh tunneling mechanism. Users cannot use Globus online and/or Cea to transfer files between new and old file systems, nor they can use Cea and Globus Online to transfer files from their local devices to Arrow's file system. However the use of ssh tunneling offers an alternative way to securely transfer files to Arrow over the Internet using ssh protocol and Chizen as a ssh server. Users are encouraged to contact HPCC for further guidance. Here is example of tunneling via Chizen:
scp -J <user_id>@chizen.csi.cuny.edu <file_to_transfer> <user_id>@arrow:/scratch/<user_id/.
Users must submit their password twice for Chizen and for Arrow.
Files are tunneled through but not copied to chizen. Note that files copied to Chizen will be removed.
Set up execution environment
Overview of LMOD environment modules system
Each of the applications, libraries and executables requires specific environment. In addition many software packages and/or system packages exist in different versions. To ensure proper environment for each and every application, library or system software CUNY-HPCC applies the environment module system which allow quick and easy way to dynamically change user's environment through modules. Each module is a file which describes needed environment for the package.Modulefiles may be shared by all users on a system and users may have their own collection of module files. Note that on old servers (Penzias, Appel) HPCC utilizes TCL based modules management system which has less capabilities than LMOD. On Arrow HPCC uses only LMOD environment. management system. The latter is Lua based and has capabilities to resolve hierarchies. It is important to mentioned that LMOD system understands and accepts the TCL modules Thus user's module existing on Appel or Penzias can be transferred and used directly on Arrow. The LMOD also allows to use shortcuts. In addition users may create collections of modules and store the later under particular name. These collections can be used for "fast load" of needed modules or to supplement or replacement of the shared modulefiles. For instance ml can be used as replacement of command module load.
Modules categories
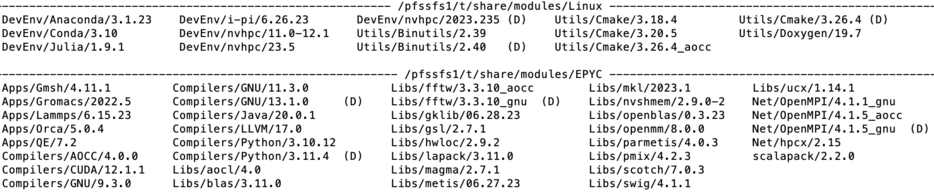
module category Library
Lmod modules are organized in categories. On Arrow the categories are Compilers, Libraries (Libs), Utilities(Util), Applications. Development Environments(DevEnv) and Communication (Net). To check content of each category the users may use the command module category <name of the category>. The picture above shows the output. In addition the version of the product is showed in module file name. Thus the line
Compilers/GNU/13.1.0
shown in EPYC directory denotes the module file for GNU (C/C++/fortran) compiler ver 13.1.0. tuned for AMD architecture.
List of available modules
To get list of available modules the users may use the command
module avail
The output of this command for Arrow server is shown.The (D) after the module's name denotes that this module is default. The (L) denotes that the module is already loaded.
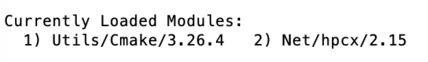
Load module(s) and check for loaded modules
Command module load <name of the module> OR module add<name of the module> loads a requested module. For example the below command load modules for utility cmake and network interface. User may check which modules are already loaded by typing module list. The figure below shows the output of this command
module load Utils/Cmake/3.26.4
module add Net/hpcx/2.15
module list
Another command which is equivalent to module load is module add as it is shown in above example.
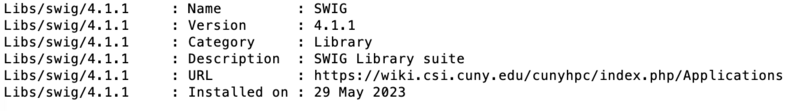
Module details
The information about module is available via whatis command for library swig:
module whatis Libs/swig
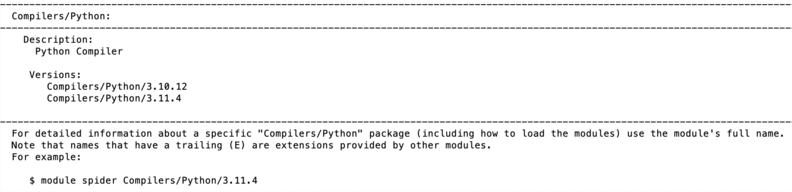
Searching for modules
Modules can be searched by module spider command. For instance the search of Python modules gives the following output:
module spider Python
t Each modulefile holds information needed to configure the shell environment for a specific software application, or to provide access to specific software tools and libraries. Modulefiles may be shared by all users on a system and users may have their own collection of module files. The users' collections may be used for "fast load" of needed modules or to supplement or replace the shared modulefiles.
Compiling user's developed codes on Arrow
Arrow login node is Intel X86_64 server with two K20m GPU. Codes can be compiled there and executable can run on AMD nodes, but with basic X86_64/AMD compatibility. For better results HPCC recommends to:
- compile codes directly on nodes where the codes will be run.
- use AMD optimized libraries such as ACML and AMD tuned compilers (AOCC). Users should read AOCC user manual for optimization options.
- the GNU compilers can be used as well but optimal performance on nodes is not guaranteed.
To compiler code directly on a node HPCC recommends the users to submit the batch job (alternative is to use interactive job - see below). Here is an example of compilation of parallel FORTRAN 77 code on a node member of particular partition.
#!/bin/bash
#SBATCH --nodes=1 # request for one node
#SBATCH --job_name=<job_name>
#SBATCH --partition=<partition where to compile> #one of the partitions when the user is registered
#SBATCH --qos=<qos for group e.g. qosmath>
#SBATCH --ntasks=1
#SBATCH --mem=64G
echo $SLURM_CPUS_PER_TASK
module purge
module load Compilers/AOCC/4.0.0. # load compiler
module load Net/OpenMPI/4.1.5_aocc # load OpenMPI library
mpif77 -o <executable> -O.. <source> #invokes compilation. Add appropriate optimization flags
Batch job submission system (SLURM)
This section below describes use of SLURM batch job submission system on Arrow. However many of examples can be also used on older servers like Penzias or Appel. Note that Penzais has outdated K20m GPU so pay attention and specify correctly the GPU type correctly in GPU constraints. SLURM is open source scheduler and batch system which is implemented at HPCC. SLURM is used on all servers to submit jobs.
SLURM script structure
A Slurm script must do three things:
- prescribe the resource requirements for the job
- set the environment
- specify the work to be carried out in the form of shell commands
The simple SLURM script is given below
#!/bin/bash
#SBATCH --job-name=test_job # some short name for a job
#SBATCH --nodes=1 # node count
#SBATCH --ntasks=1 # total number of tasks across all nodes
#SBATCH --cpus-per-task=1 # cpu-cores per task (>1 if multi-threaded tasks)
#SBATCH --mem-per-cpu=16 # memory per cpu-core
#SBATCH --time=00:10:00 # total run time limit (HH:MM:SS)
#SBATCH --mail-type=begin # send email when job begins
#SBATCH --mail-type=end # send email when job ends
#SBATCH --mail-user=<valid user email>
cd $SLURM_SUBMIT_DIR # change to directory from where jobs starts
The first line of a Slurm script above specifies the Linux/Unix shell to be used. This is followed by a series of #SBATCH directives which set the resource requirements and other parameters of the job. The script above requests 1 CPU-core and 4 GB of memory for 1 minute of run time. Note that #SBATCH is command to SLURM while the # not followed by SBATH is interpret as comment line. Users can submit 2 types of jobs - batch jobs and interactive jobs:
sbatch <name-of-slurm-script> submits job to the scheduler
salloc requests an interactive job on compute node(s) (see below)
Job(s) execution time
The job execution time is sum with them the job waits in SLURM partition (queue) before being executed on node(s) and actual running time on node. For the parallel corder the partition time (time job waits in partition) increases with increasing resources such as number of CPU-cores. On other hand the execution time (time on nodes) decreases with inverse of resources. Each job has its own "sweet spot" which minimizes the time to solution. Users are encouraged to run several test runs and to figure out what amount of asked resources works best for their job(s).
Partitions and quality of service (QOS)
In SLURM terminology partition has a meaning of a queue. Jobs are placed in partition for execution. QOS mechanism to apply policies and thus to control the resources on user lever or on partition level. In particular when applied to partition the QOS allows to create a 'floating' partition - namely a partition that gets all assigned resources (nodes) but allows to run on the number of resources in it. HPCC uses QOS on all partitions on Arrow to set policies on these partitions. Thus the HPCC ensures fair share policy for resources in each partition and controls access to the partitions according to user's status. For instance the QOS establishes that only core members of the NSF grant have access to NSF funded resources listed in partition partnsf (see below) . Currently the partitions and QOS on Arrow are:
| Partition | Nodes | QOS | Allowed Users | Partition limitations | Notes |
|---|---|---|---|---|---|
| partnsf | n130,n131 | qosnsf | All registered core participants of NSF grant | max 128 cores /240h per job | only core participants of NSF grant OAC-2215760 |
| partchem | n133, n136 | qoschem | All registered users from Prof. S.Loverde Group and ASRC | no limits | privare partition |
| partmath | n138,n137 | qosmath | All registered users from Prof. A.Poje and Prof. A.Kuklov Groups | no limits | privare partition |
| partcfd | n137 | high | All registered users from Prof. A Poje Group | no limits | privare partition |
| parthphys | n138 | high | All registered users from Prof. Kuklov Group | no limits | privare partition |
| partsym | n133 | qossymhigh | All registered users from Prof. Loverde Group | no limits | privare partition |
| partasrc | n136 | qosacrchigh | All registered users from ASRC | no limits | privare partition |
Note that users can submit job only to partition they are registered to e.g. jobs from users registered to partnsf will be rejected on other partitions (and vice versa). The nodes n130 and n131 are accessible only for For instance the core user A from Sharon Loverde Group can use partnsf and QOS qosnsf to submit jobs on
Working with QOS and partitions on Arrow
Every job submission script on Arrow must hold proper description of QOS and partition. For instance all jobs intended to use node n133 must have the following lines:
#SBATCH --qos=qoschem
#SBATCH --partition partchem
In similar way all jobs intended to use n130 and n131 must have in their job submission script:
#SBATCH --qos=qosnsf
#SBATCH --partition partnsf
Note that Penzias do not use QOS. Thus users must adapt scripts they copy from Penzias server to match QOS requirements on Arrow.
Submitting serial (sequential jobs)
These jobs utilize only a single CPU-core. Below is a sample Slurm script for a serial job in partition partchem. Users must add lines for QOS and partition as was explained above. :
#!/bin/bash
#SBATCH --job-name=serial_job # short name for job
#SBATCH --nodes=1 # node count always 1
#SBATCH --ntasks=1 # total number of tasks aways 1
#SBATCH --cpus-per-task=1 # cpu-cores per task (>1 if multi-threaded tasks)
#SBATCH --mem-per-cpu=8G # memory per cpu-core
#SBATCH --qos=qoschem
#SBATCH --partition partchem
cd $SLURM_SUBMIT_DIR
srun ./myjob
In above script requested resources are:
- --nodes=1 - specify one node
- --ntasks=1 - claim one task (by default 1 per CPU-core)
Job can be submitted for execution with command:
sbatch <name of the SLURM script>
For instance if the above script is saved in file named serial_j.sh the command will be:
sbatch serial_j.sh
Submitting multithread job
Some software like MATLAB or GROMACS are able to use multiple CPU-cores using shared-memory parallel programming models like OpenMP, pthreads or Intel Threading Building Blocks (TBB). OpenMP programs, for instance, run as multiple "threads" on a single node with each thread using one CPU-core. The example below show how run thread-parallel on Arrow. Users must add lines for QOS and partition as was explained above.
#!/bin/bash
#SBATCH --job-name=multithread # create a short name for your job
#SBATCH --nodes=1 # node count
#SBATCH --ntasks=1 # total number of tasks across all nodes
#SBATCH --cpus-per-task=4 # cpu-cores per task (>1 if multi-threaded tasks)
#SBATCH --mem-per-cpu=4G # memory per cpu-core
export OMP_NUM_THREADS=$SLURM_CPUS_PER_TASK
In this script the the cpus-per-task is mandatory so SLURM can run the multithreaded task using four CPU-cores. Correct choice of cpus-per-task is very important because typically the increase of this parameter decreases in execution time but increases waiting time in partition(queue). In addition these type of jobs rarely scale well beyond 16 cores. However the optimal value of cpus-per-task must be determined empirically by conducting several test runs. It is important to remember that the code must be 1. muttered code and 2. be compiled with multithread option for instance -fomp flag in GNU compiler.
Submitting distributed parallel job
These jobs use Message Passing Interface to realize the distributed-memory parallelism across several nodes. The script below demonstrates how to run MPI parallel job on Arrow. Users must add lines for QOS and partition as was explained above.
#!/bin/bash
#SBATCH --job-name=MPI_job # short name for job
#SBATCH --nodes=2 # node count
#SBATCH --ntasks-per-node=32 # number of tasks per node
#SBATCH --cpus-per-task=1 # cpu-cores per task (>1 if multi-threaded tasks)
#SBATCH --mem-per-cpu=16G # memory per cpu-core
cd $SLURM_SUBMIT_DIR
srun ./mycode <args>. # mycode is in local directory. For other places provider full path
The above script can be easily modified for hybrid (OpenMP+MPI) by changing the cpu-per-task parameter. The optimal value of --nodes and --ntasks for a given code must be determined empirically with several test runs. In order to run decrease communication the users shown try to run large jobs by taking the whole node rather than 2 chunks from 2 (or more nodes). In addition for large memory jonbs users must use --mem rather than mem-per-cpu. Below is an SLURM script example for submission of large memory MPI job with 128 cores on a single mode. Obviously is better this type of job to be run on single node rather than on two times 64 cores from 2 nodes. To achieve that users may use the following SLURM prototype script:
#!/bin/bash
#SBATCH --job-name MPI_J_2
#SBATCH --nodes 1
#SBATCH --ntasks 128 # total number of tasks
#SBATCH --mem 40G # total memory per job
#SBATCH --qos=qoschem
#SBATCH --partition partchem
cd $SLURM_SUBMIT_DIR
srun ...
In above script the requested resources are 128 cores on one node. Note that unused memory on this node will not be accessible to other jobs. In difference to previous script the memory is referred as total memory for a job via parameter --mem.
Submitting Hybrid (OMP+MPI) job on Arrow
#!/bin/bash
#SBATCH --job-name=OMP_MPI # name of the job
#SBATCH --ntasks=24 # total number of tasks aka total # of MPI processes
#SBATCH --nodes=2 # total number of nodes
#SBATCH --tasks-per-node=12 # number of tasks per node
#SBATCH --cpus-per-task=2 # number of OMP threads per MPI process
#SBATCH --mem-per-cpu=16G # memory per cpu-core
#SBATCH --partition=partnsf
#SBATCH --qos=qosnsf
cd $SLURM_SUBMIT_DIR
export OMP_NUM_THREADS=$SLURM_CPUS_PER_TASK
export SRUN_CPUS_PER_TASK=$SLURM_CPUS_PER_TASK
srun ...
The above script si prototype and shows how to allocate 24 MPI treads with 12 cores per node. Each MPI tread initiates 2 OMP threads. For actual working script users must add QOS and partition information and adjust their requirements for the memory.
GPU jobs
On arrow each of the nodes has 8 GPU A40 with 80GB on board. To use GPUs in a job users must add the --gres option in SBATH line for cpu resources. The example below demonstrates a GLU enabled SLURM script. Users must add lines for QOS and partition as was explained above.
#!/bin/bash
#SBATCH --job-name=GPU_J # short name for job
#SBATCH --nodes=1 # number of nodes
#SBATCH --ntasks=1 # total number of tasks across all nodes
#SBATCH --cpus-per-task=1 # cpu-cores per task (>1 if multi-threaded tasks)
#SBATCH --mem-per-cpu=16G # memory per cpu-core
#SBATCH --gres=gpu:1 # number of gpus per node max 8 for Arrow
cd $SLURM_SUBMIT_DIR
srun ... <code> <args>
GPU constrains
On Appel the nodes in partnsf, partchem and partmath have different GPU types (A30, A40 and A100). The type of GPU can be specified in SLURM by use constraint on the GPU SKU, GPU generation, or GPU compute capability. Here is example:
#SBATCH --gres=gpu:1 --constraint='gpu_sku:V100' # allocates one V100 GPU
#SBATCH --gres=gpu:1 --constraint='gpu_gen:Ampere' # allocates one Ampere GPU (A40 or A100)
#SBATCH --gres=gpu:1 --constraint='gpu_cc:12.0' # allocates GPU per computability (generation)
#SBATCH --gres=gpu:1 --constraint='gpu_mem:32GB' # allocates GPU with 32GB memory on board
#SBATCH --gres=gpu:1 --constraint='nvlink:2.0'. # allocates GPU linked with NVLink
Parametric jobs via Job Array
Job arrays are used for running the same job multiple times but with only slightly different parameters. The below script demonstrates how to run such a job on Arrow. Users must add lines for QOS and partition as was explained above. NB! The array numbers must be less than the maximum number of jobs allowed in the array.
#!/bin/bash
#SBATCH --job-name=Array_J # short name for job
#SBATCH --nodes=1 # node count
#SBATCH --ntasks=1 # total number of tasks across all nodes
#SBATCH --cpus-per-task=1 # cpu-cores per task (>1 if multi-threaded tasks)
#SBATCH --mem-per-cpu=16G # memory per cpu-core
#SBATCH --output=slurm-%A.%a.out # stdout file (standart out)
#SBATCH --error=slurm-%A.%a.err # stderr file (standart error)
#SBATCH --array=0-3 # job array indexes 0, 1, 2, 3
cd $SLURM_SUBMIT_DIR
<executable>
Interactive jobs
These jobs are useful in development or test phase and rarely are required in a workflow. It is not recommend to use interactive jobs as main type of jiobs since they consume more resources that regular batch jobs. To set up interactive job the users first have to 1. start interactive shell and 2 "reserve" the resources. teh example below describes that.
srun -p interactive --pty /bin/bash # starts interactive session
Once the interactive session is running the users must "reserve" resources needed for actual job:
salloc --ntasks=8 --ntasks-per-node=1 --cpus-per-task=2. # allocates resources
salloc: CPU resource required, checking settings/requirements...
salloc: Granted job allocation ....
salloc: Waiting for resource configuration
salloc: Nodes ... # system reports back where the resources were allocated